NucTools – software package for the analysis of chromatin feature occupancy profiles from high-throughput sequencing data.
NucTools is a suite of Perl scripts as well as MATLAB- and R-based visualization programs for a nucleosome-centred downstream analysis of deep sequencing data. NucTools accounts for the continuous distribution of nucleosome occupancy. It allows calculations of nucleosome occupancy profiles averaged over several replicates, comparisons of nucleosome occupancy landscapes between different experimental conditions, and the estimation of the changes of integral chromatin properties such as the nucleosome repeat length. Furthermore, NucTools facilitates the annotation of nucleosome occupancy with other chromatin features like binding of transcription factors or architectural proteins, and epigenetic marks like histone modifications or DNA methylation.
Citation:
Vainshtein Y., Rippe K. and Teif V.B. (2017). NucTools: analysis of chromatin feature occupancy profiles from high-throughput sequencing data. BMC Genomics 18, 158 | Open access article
Developers: Yevhen Vainshtein and Vladimir B. Teif
Support: yevhen.vainshtein AT igb.fraunhofer.de
Download: The latest version and description is available at GitHub
Typical workflow:
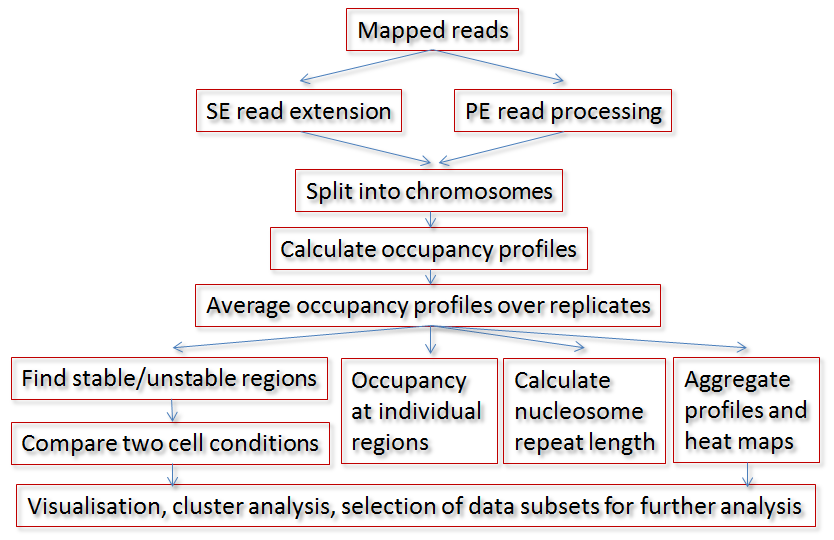
NucTools is composed of Perl and R scripts, as well as a standalone program Cluster Map Builder (CMB) to visualise chromatin heatmaps and perform cluster analysis:

The Cluster Map Builder has a simple graphical user interface exemplified below, which is described in the user manual.
![]()
The latest version of NucTools is available at GitHub
Previous versions:
NucTools 1.0 Perl scripts: NucTools_1.0.zip
NucTols 1.0 (outdated) versions of Matlab scripts: ClusterMaps_Builder_src.zip
![]() Our software NucTools is included as part of NucPosDB, a manually curated database of experimental nucleosome maps in vivo, cfDNA and computational tools related to nucleosome positioning, nucleosome-scale 3D genome organisation and cfDNA nucleosomics.
Our software NucTools is included as part of NucPosDB, a manually curated database of experimental nucleosome maps in vivo, cfDNA and computational tools related to nucleosome positioning, nucleosome-scale 3D genome organisation and cfDNA nucleosomics.
Jump to: NucPosDB front page | experimental nucleosome maps in vivo | experimental cfDNA datasets | tools for analysis of nucleosome mapping experiments | tools for prediction of nucleosome maps from DNA sequence| tools for analysis of sequenced cfDNA